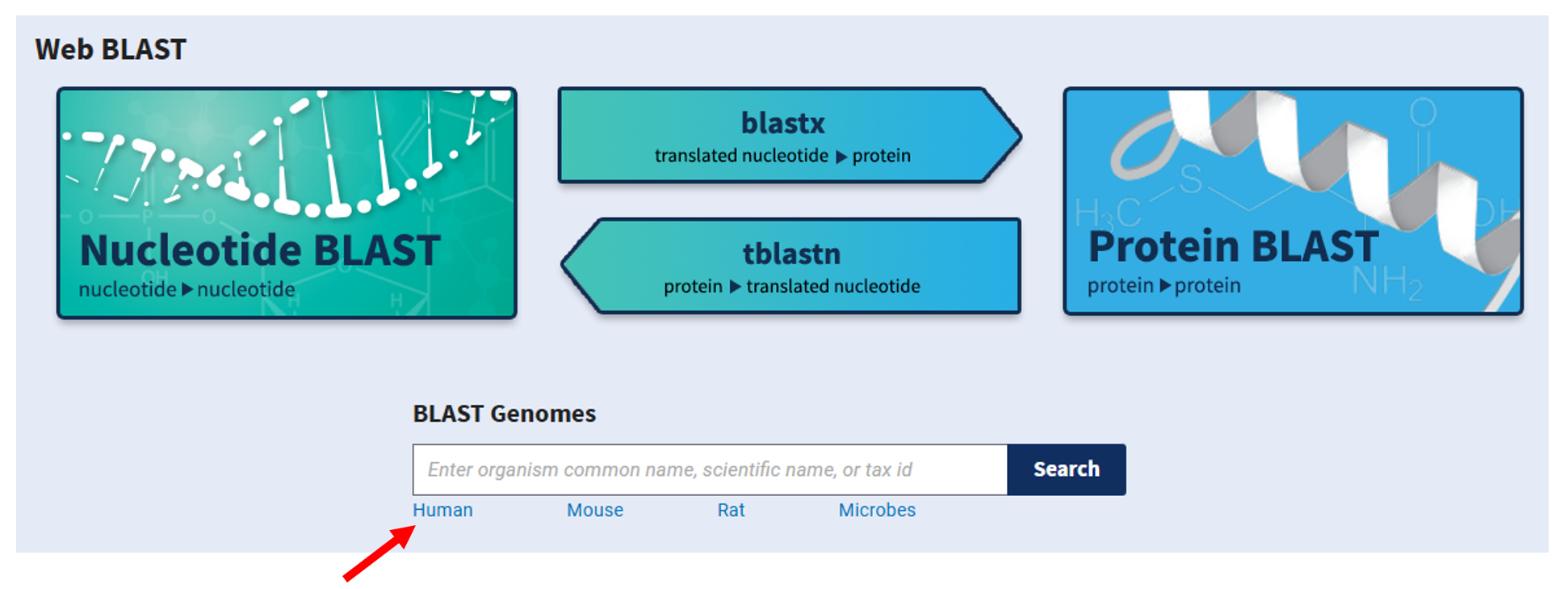
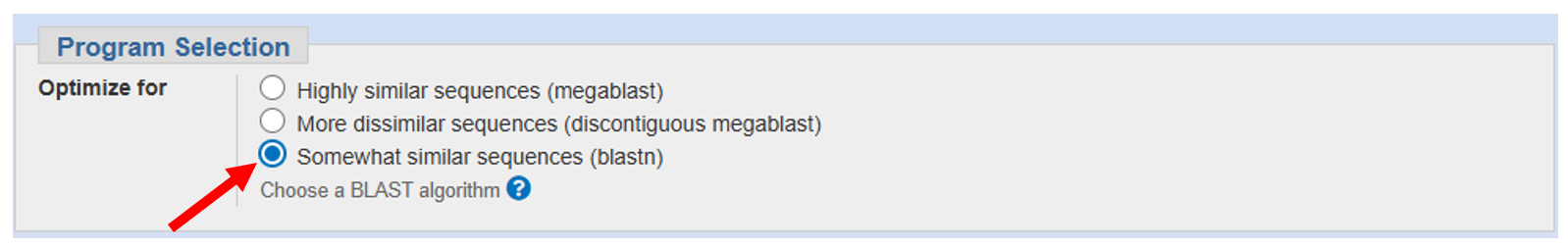
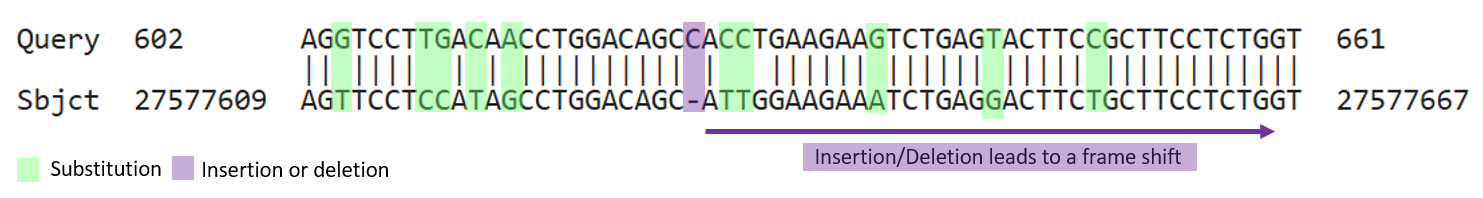
To discover the role of this mysterious sequence, we will search for proteins in a database that show high similarity to a translation of Sequence R.
To do this we will use a sequence search engine, BLAST, to search sequences in the database at the National Center for Biotechnology Information (NCBI).